Events
Include expired: true
and Keywords: eLearning or Genome Mapping or SystemsBiology or Workflows or Nanotechnology or Medicine or Docker
and Scientific topics: Informatics or Bioinformatics
TeSS makes use of some necessary cookies to provide its core functionality. Additionally, we make use of Google Analytics to discover how people are using TeSS in order to help us improve the service. To opt out of this, choose the "Allow necessary cookies" option.
See our Privacy Policy for more information.
You can modify your cookie preferences at any time here, or from the link in the footer.
Include expired: true
and Keywords: eLearning or Genome Mapping or SystemsBiology or Workflows or Nanotechnology or Medicine or Docker
and Scientific topics: Informatics or Bioinformatics
In the left-hand column of the Google Calendar main view, click the arrow to the right of "Other calendars" and click "Add by URL". In the form that appears, paste in the URL from the box above, and click the button to confirm.
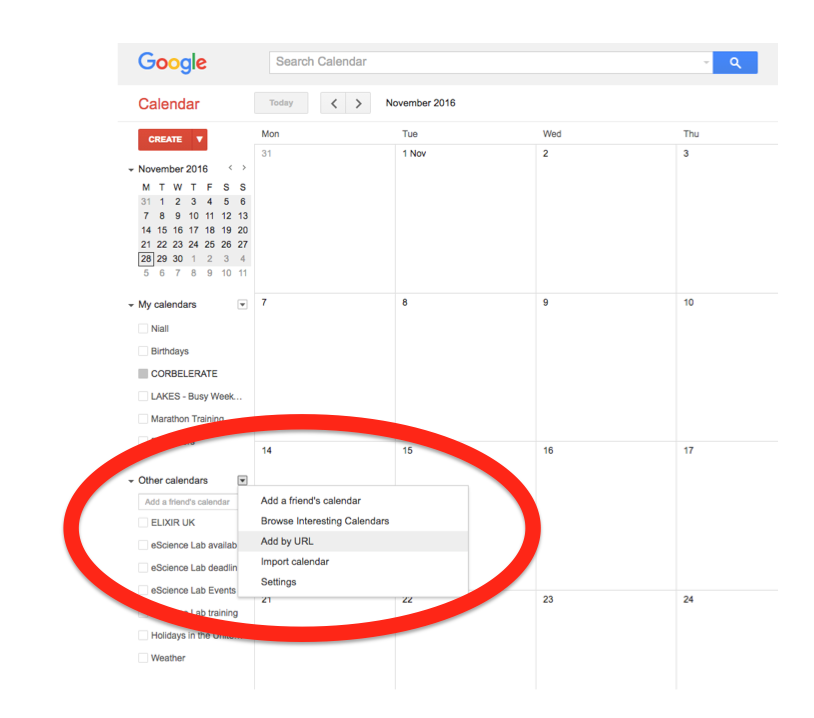
Please note, it may take a while for newly created events in TeSS to synchronise with your Google Calendar.
12 - 15 November 2016
Heidelberg, Germany
Face-to-face
Bioinformatics Genomics Systems biology SystemsBiology OtherEvents ISCB24 - 28 April 2017
Edinburgh, United Kingdom
Face-to-face
Bioinformatics Linux Workflows11 May 2017 @ 12:00 - 13:00
University of Melbourne, Australia
Face-to-face
Bioinformatics programming Workflows21 November 2017 @ 09:00 - 17:00
Oeiras, Portugal
Face-to-face
Bioinformatics Teaching eLearning EeLP bioinformatics28 February 2018 @ 14:00 - 15:00
Online
Workflows Genomics Bioinformatics CWL HPC LSF21 March 2018 @ 13:30 - 16:30
Carlton, Australia
Face-to-face
Bioinformatics Docker14 - 18 May 2018
Edinburgh, United Kingdom
Face-to-face
Bioinformatics Workflows Linux21 June 2018 @ 13:30 - 15:30
Carlton, Australia
Face-to-face
Bioinformatics bioinformatics Docker25 - 26 June 2018
Torino, Italy
Face-to-face
Bioinformatics Computer science Docker Reproducibility Container computer-science20 August 2018 @ 15:00 - 16:30
Online
Bioinformatics GWAS study Workflows Computational biology Genotyping experiment Population genomics Nextflow Docker H3ABioNet GWAS Genotyping array analysis bioinformatics Africa Population Genomics Reproducible Science H3Africa genotyping array High performance computing Cloud computing GWAS workflow H3ABioNet GWAS 2018 Lecture Series
Note, this map only displays events that have geolocation information in
TeSS.
For the complete list of events in TeSS, click the grid tab.