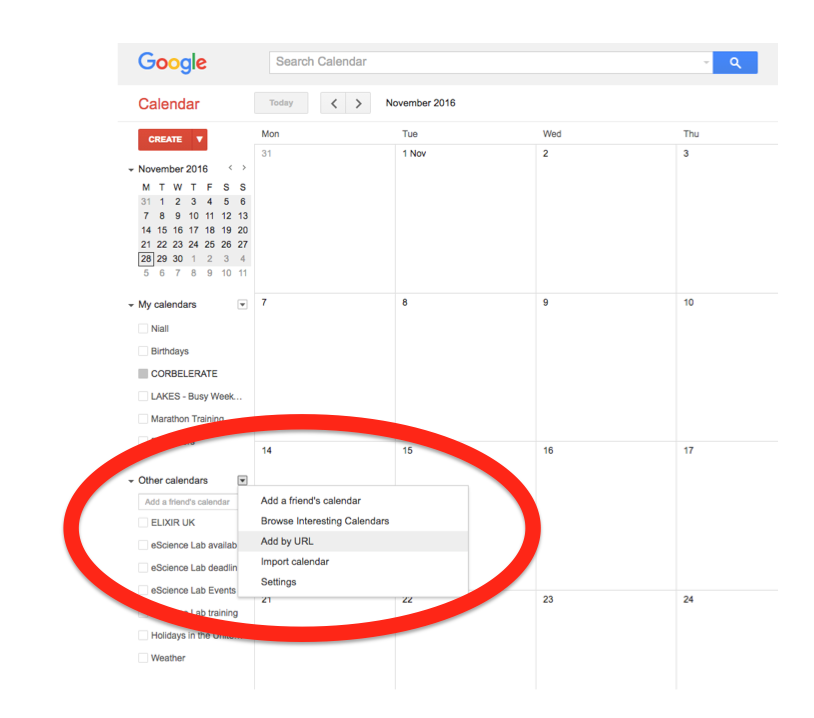
Events
Include expired: true
and Content provider: ELIXIR Norway or ELIXIR Portugal or EMBL-ABR
and Keywords: eLearning or Immunity or EeLP or Unix or Computer Programming or Model-based Population Genetics
- 1
- 2
Note, this map only displays events that have geolocation information in
TeSS.
For the complete list of events in TeSS, click the grid tab.