Events
City: Bogota or Heidelberg or Hinxton or Leuven or Amsterdam UMC (location AMC) or Leiden University or Cambridge
TeSS makes use of some necessary cookies to provide its core functionality. Additionally, we make use of Google Analytics to discover how people are using TeSS in order to help us improve the service. To opt out of this, choose the "Allow necessary cookies" option.
See our Privacy Policy for more information.
You can modify your cookie preferences at any time here, or from the link in the footer.
City: Bogota or Heidelberg or Hinxton or Leuven or Amsterdam UMC (location AMC) or Leiden University or Cambridge
In the left-hand column of the Google Calendar main view, click the arrow to the right of "Other calendars" and click "Add by URL". In the form that appears, paste in the URL from the box above, and click the button to confirm.
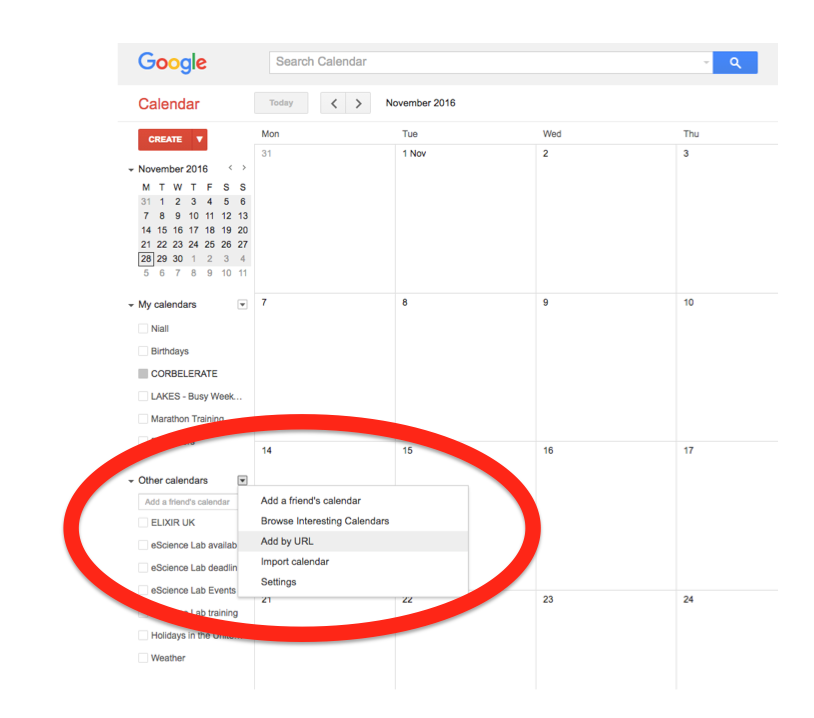
Please note, it may take a while for newly created events in TeSS to synchronise with your Google Calendar.
11 January - 8 May 2024
Leuven, Belgium
Face-to-face
career development29 January - 29 April 2024
Leuven, Belgium
Face-to-face
leadership & management15 April - 7 May 2024
Leuven, Belgium
Face-to-face
communication skills19 April - 2 May 2024
Leuven, Belgium
Face-to-face
advanced bioinformatics omics29 - 30 April 2024
Cambridge, United Kingdom
Face-to-face
HDRUK1 - 2 May 2024
Cambridge, United Kingdom
Face-to-face
HDRUK8 May 2024
Leuven, Belgium
Face-to-face
leadership & management personal effectiveness career development13 - 24 May 2024
Leuven, Belgium
Face-to-face
communication skills13 - 15 May 2024
Cambridge, United Kingdom
Face-to-face
HDRUK16 - 20 May 2024
Cambridge, United Kingdom
Face-to-face
Bioinformatics Data mining Data visualisation Functional genomics Transcriptomics HDRUK
Note, this map only displays events that have geolocation information in
TeSS.
For the complete list of events in TeSS, click the grid tab.