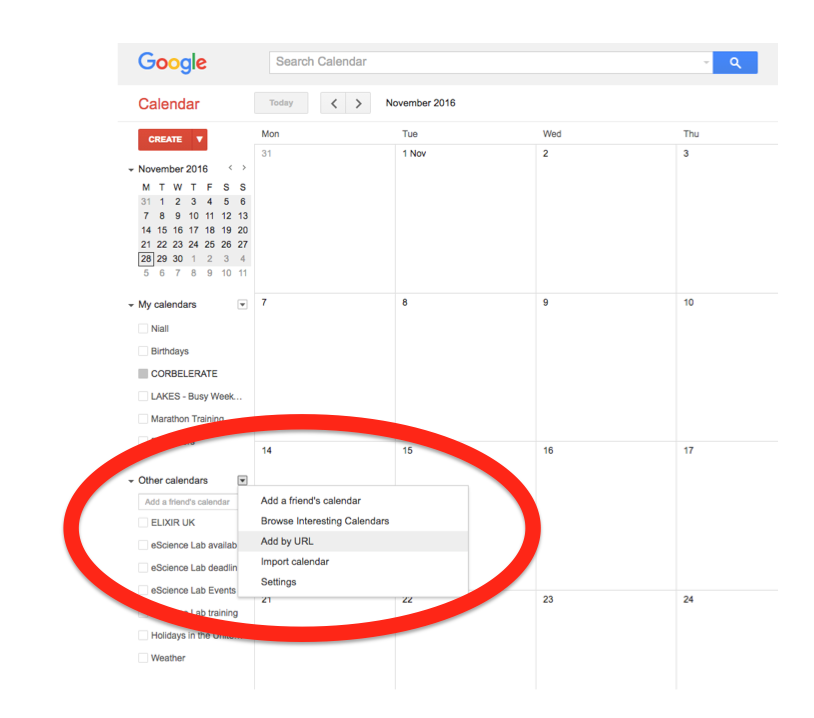
Events
Include expired: true
and City: Bogota or Cambridge or Buenos Aires
and Target audience: Postdocs and Staff members from the University... or and industrial investigations into diagnostic a... or The course is aimed primarily at mid-career sci...