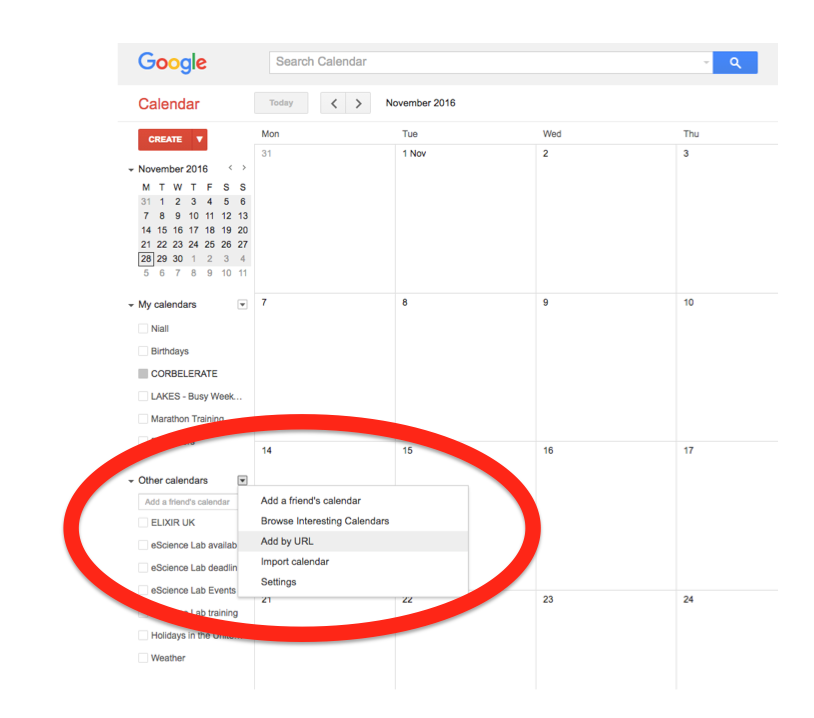
Events
Include expired: true
and City: Bogota or Bern or Cambridge or Faro
and Target audience: Industry or This workshop is aimed at researchers who want ... or <span style="color:#0000FF">All Members of the ... or The module is suitable for researchers interest... or The course is aimed at <b>bench biologists and ... or view